The High-Throughput Toolkit (httk)¶
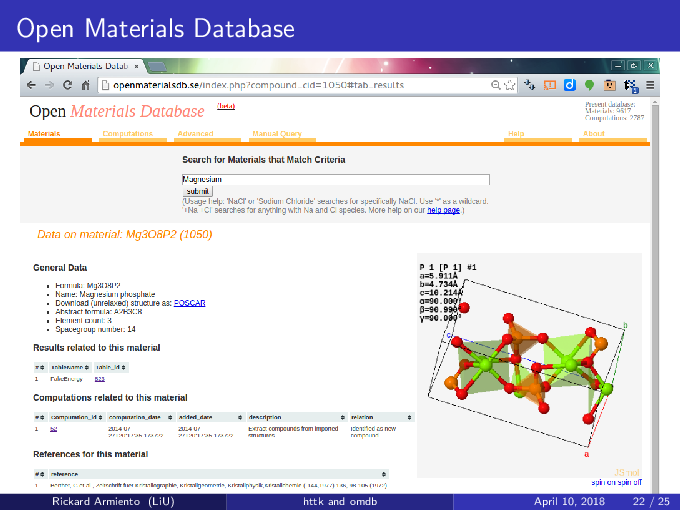
This website documents the High-Throughput Toolkit (httk). Looking for the Open Materials Database? It is at: http://openmaterialsdb.se
About the High-Throughput Toolkit¶
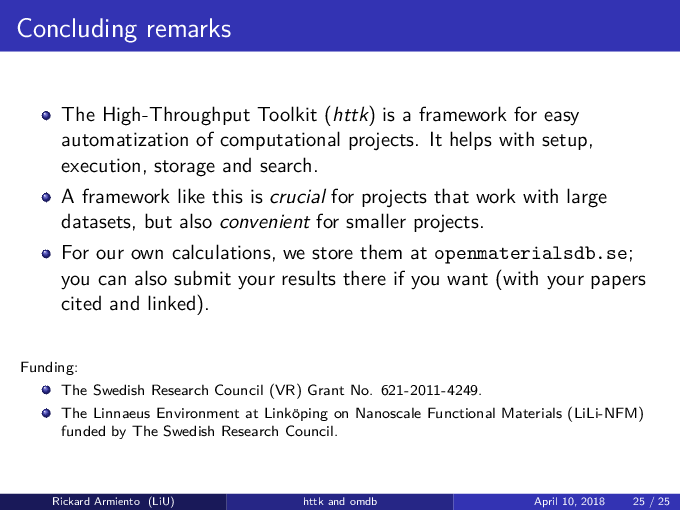
The High-Throughput Toolkit (httk) is a toolkit for:
- Preparing and running calculations.
- Analyzing the results.
- Store the results and outcome in a global and/or in a personalized database.
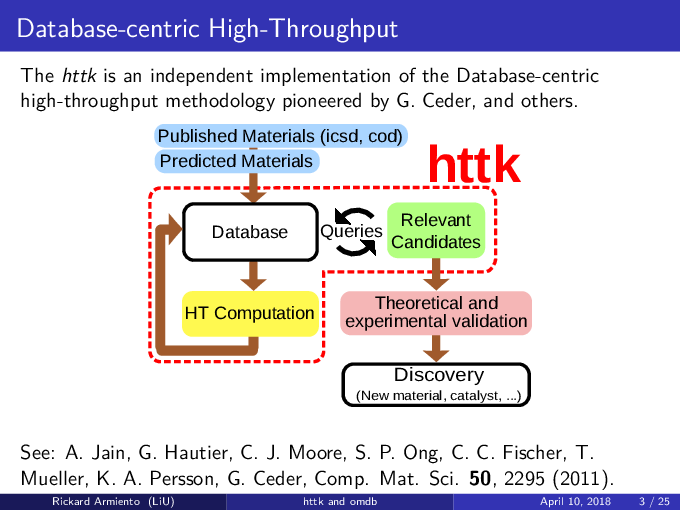
httk is an independent implementation of the database-centric high-throughput methodology pioneered by Ceder et al., and others. [see, e.g., Comp. Mat. Sci. 50, 2295 (2011)]. httk is presently targeted at atomistic calculations in materials science and electronic structure, but aims to be extended into a library useful also outside those areas.
Quickstart¶
Httk presently consists of a python library and a few programs. If you just want access to use (rather than develop) the python library, and do not need the external programs, the install is very easy.
(Note: for httk version 2.0 we will go over to a single ‘script’ endpoint,
httk, for which the pip install step should be sufficient to get a full install.)
Install to access just the python library¶
You need Python 2.7 and access to pip in your terminal window. (You can get Python and pip, e.g., by installing the Python 2.7 version of Anaconda, https://www.anaconda.com/download, which should give you all you need on Linux, macOS and Windows.)
Issue in your terminal window:
pip install httk
If you at a later point want to upgrade your installation, just issue:
pip install httk --upgrade
You should now be able to simply do import httk in your python programs to use the httk python library.
Alternative install: python library + binaries + ability to develop httk¶
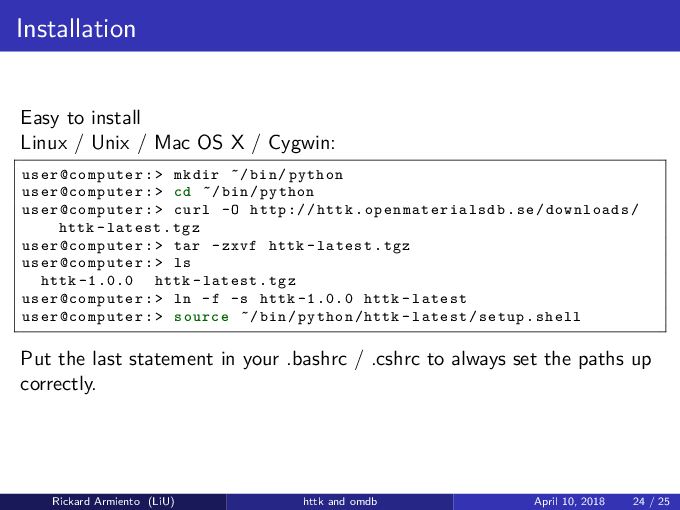
In addition to Python 2.7 and pip, you also need git. You can get git from here: https://git-scm.com/
Issue in your terminal window:
git clone https://github.com/rartino/httk cd httk pip install --editable . --user
If you at a later point want to upgrade your installation, just go back to the httk directory and issue:
git pull pip install . --upgrade --user
To setup the paths to the httk programs you also need to run:
source /path/to/httk/init.shell
where
/path/to/httkshould be the path to where you downloaded httk in the steps above. To make this permanent, please add this line to your shell initialization script, e.g., ~/.bashrc
You are now ready to use httk.
Notes:
The above instructions give you access to the latest stable release of httk. To get the latest developer relase (which may or may not work), issue:
git checkout devel pip install . --upgrade --userin your httk directory. To switch back to the stable release, do:
git checkout master pip install . --upgrade --userAn alternative to installing with
pip installis to just run httk out of the httk directory. In that case, skip the pip install step above and just appendsource ~/path/to/httk/init.shellto your shell init files, with~/path/to/httkreplaced by the path of your httk directory.)*
A few simple usage examples¶
Load a cif file or poscar¶
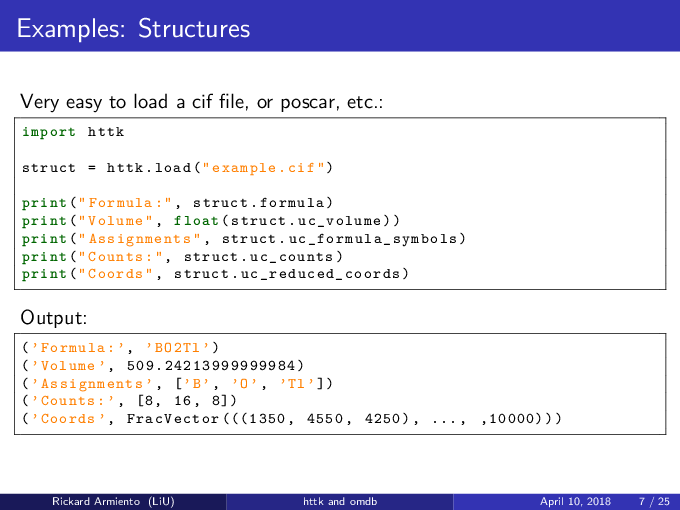
This is a very simple example of just loading a structure from a .cif file and writing out some information about it.
import httk
struct = httk.load("example.cif")
print("Formula:", struct.formula)
print("Volume:", float(struct.uc_volume))
print("Assignments:", struct.uc_formula_symbols)
print("Counts:", struct.uc_counts )
print("Coords:", struct.uc_reduced_coords)
Running this generates the output:
('Formula:', 'BO2Tl')
('Volume', 509.24213999999984)
('Assignments',['B', 'O', 'Tl'])
('Counts:', [8, 16, 8])
('Coords', FracVector(((1350,4550,4250) , ... , ,10000)))
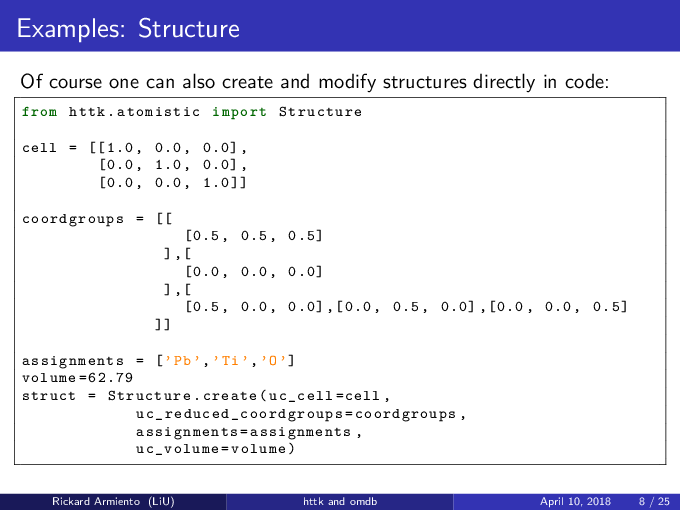
Create structures in code¶
from httk.atomistic import Structure
cell = [[1.0, 0.0, 0.0] ,
[0.0, 1.0, 0.0] ,
[0.0, 0.0, 1.0]]
coordgroups = [[
[0.5, 0.5, 0.5]
],[
[0.0, 0.0, 0.0]
],[
[0.5, 0.0, 0.0], [0.0, 0.5, 0.0], [0.0, 0.0, 0.5]
]]
assignments = ['Pb' ,'Ti' ,'O']
volume =62.79
struct = Structure.create(uc_cell = cell,
uc_reduced_coordgroups = coordgroups,
assignments = assignments,
uc_volume = volume)
Create database file, store a structure in it, and retrive it¶
import httk, httk.db
from httk.atomistic import Structure
backend = httk.db.backend.Sqlite('example.sqlite')
store = httk.db.store.SqlStore(backend)
tablesalt = httk.load('NaCl.cif')
store.save(tablesalt)
arsenic = httk.load('As.cif')
store.save(arsenic)
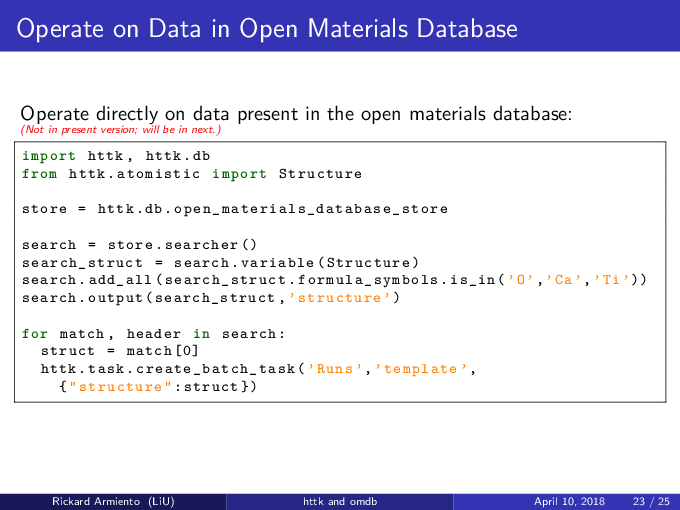
# Search for anything with Na
search = store.searcher()
search_struct = search.variable(Structure)
search.add(search_struct.formula_symbols.is_in('Na'))
search.output(search_struct, 'structure')
for match, header in list(search):
struct = match[0]
print "Found structure", struct.formula, [str(struct.get_tags()[x]) for x in struct.get_tags()]
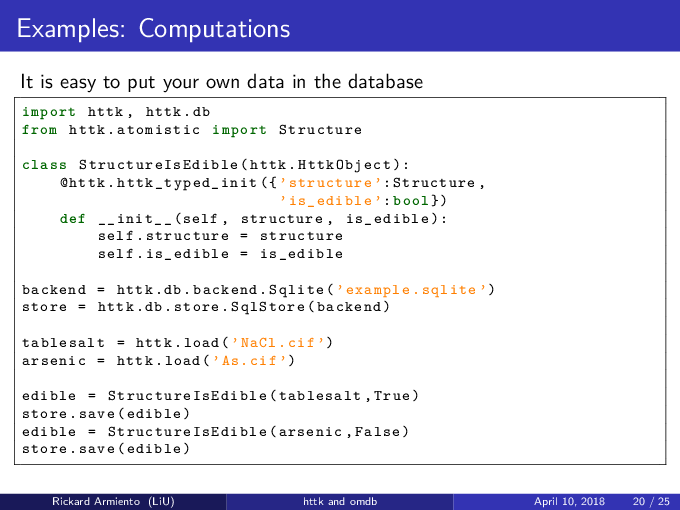
Create database file and store your own data in it¶
#!/usr/bin/env python
import httk, httk.db
from httk.atomistic import Structure
class StructureIsEdible(httk.HttkObject):
@httk.httk_typed_init({'structure': Structure, 'is_edible': bool})
def __init__(self, structure, is_edible):
self.structure = structure
self.is_edible = is_edible
backend = httk.db.backend.Sqlite('example.sqlite')
store = httk.db.store.SqlStore(backend)
tablesalt = httk.load('NaCl.cif')
edible = StructureIsEdible(tablesalt, True)
store.save(edible)
arsenic = httk.load('As.cif')
edible = StructureIsEdible(arsenic, False)
store.save(edible)
Tutorial¶
Under Tutorial/Step1, 2, ... in your httk directory you find a series of code snippets to run to see httk in action.
You can either just execute them there, or try them out in, e.g., a Jupyter notebook.
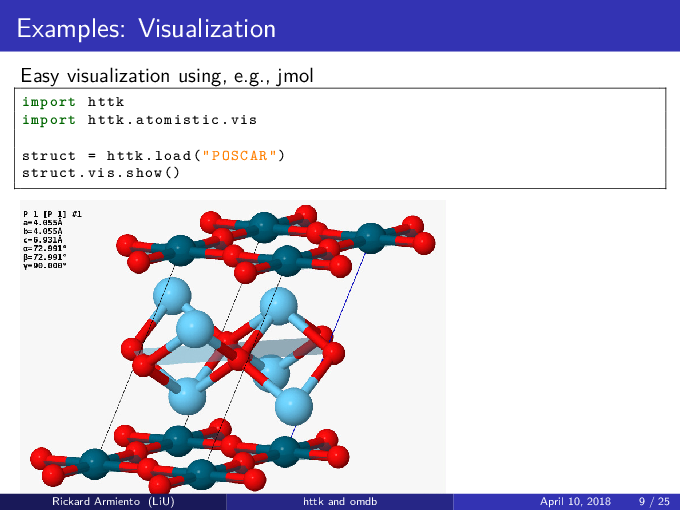
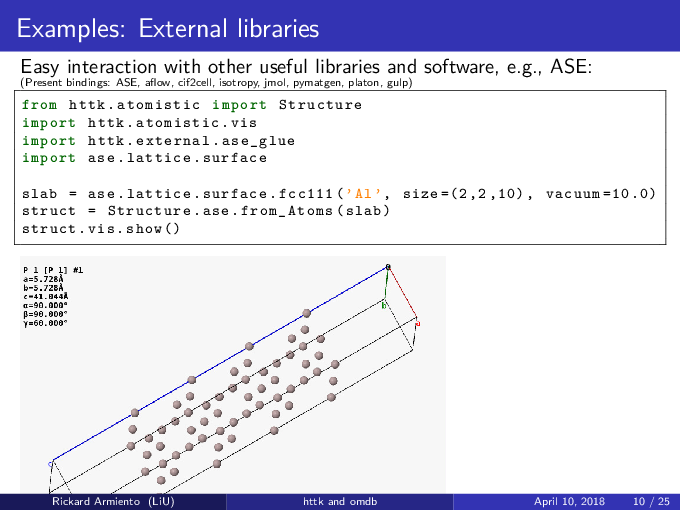
In addition to the Tutorial, there is a lot of straightforward examples of various things that can be done with httk
in the Examples subdirectory. Check the source files for information about what the various examples does.
Reporting bugs¶
We track our bugs using the issue tracker at github. If you find a bug, please search to see if someone else has reported it here:
If you cannot find it already reported, please click the ‘new issue’ button and report the bug.
Citing httk in scientific works¶
This is presently the preferred citation to the httk framework itself:
The High-Throughput Toolkit (httk), R. Armiento et al., http://httk.openmaterialsdb.se/.
Since httk can call upon many other pieces of software quite transparently, it may not be initially obvious what other software should be cited. Unless configured otherwise, httk prints out a list of citations when the program ends. You should take note of those citations and include them in your publications if relevant.
More info and help¶
For more details on installation options refer to httk Installation Instructions.
User’s guide: see httk Users’ Guide.
Workflows: for more details on how high-throughput computational workflows are executed via the runmanager.sh program, see httk Runmanager Details. This may be useful if you plan to design your own workflows using httk.
Developing / contributing to httk: see httk Developers’ Guide
For API documentation, see: outline of the httk API.Contributors¶
For a more complete list of contributors and contributions, see httk Contributors.
Acknowledgements¶
- httk has kindly been funded in part by:
- The Swedish Research Council (VR) Grant No. 621-2011-4249
- The Linnaeus Environment at Linköping on Nanoscale Functional Materials (LiLi-NFM) funded by the Swedish Research Council (VR).
License and redistribution¶
The High-Throughput Toolkit uses the GNU Affero General Public License, which is an open source license that allows redistribution and re-use if the license requirements are met. (Note that this license contains clauses that are not in the GNU Public License, and source code from httk thus cannot be imported into GPL licensed projects.)
The full license text is present in httk license.
Contact¶
Our primary point of contact is email to: httk [at] openmaterialsdb.se (where [at] is replaced by @)